 |
Protein expression in bacteria, baculovirus, or mammalian expression system |
|
 |
High-throughput, automatic and informative screen on expression level, protein aggregation state and stability |
 |
Customized assay on enzymatic activity, ligand binding, and protein-protein interactions |
 |
Different conditions are screened, monitored and imaged to determine the optimized conditions for crystallization. |
 |
High precision screening and imaging in protein structure determination |
Features and Benefits
- Versatile over-expression system featured by multiple platforms for different proteins of interest
- E. coli system for soluble protein with high stability
- Insect cell system to meet the demand for economic and large amount of eukaryotic proteins in their native conformation
- HEK system for high-quality human proteins with complicate glycosylation and other post-translational modification
- Fast, reliable and robust purification using affinity chromatography method
- Classic ploy-Histidine tag for most protein of interest
- High affinity FLAG tag for protein with very low expression level
- Alternative purification tags on customer’s request
- Fluorescence-monitored Size Exclusion Chromatography (FSEC) enables a high-throughput, automatic and informative screen on expression level, protein aggregation state and stability.
Frequently Asked Questions: Protein expression, purification and crystallization
1. Methods to study protein structures
There are three main methods: X-ray (85% of atomic structures in PDB were determined by X-ray crystallography), NMR and 3D modelling.
2. Why choose protein crystallography
The protein crystallography allows researchers to obtain an atomic resolution structure. The methods yields the correct atomic structure in solution because the structure in crystal is the same as in solution. Atomic structure contains a large amount of data compared to any other
biochemical methods.
3. Time to finish the protein crystallography
The cloning and purification will take 1-6 months depending on the protein structure. It will take 1-12 months to get a correct crystallization. The data collection will take a few weeks to 1 month. The last step of phasing and structure solution will take about 1-3 months.
4. Find best conditions for crystallization
The screening process is labor-intensive. Trial and error are recommended for different precipitants, pH and other conditions. Automation is recommended to try 100-1000 different conditions. Large crystals are helpful for final data collection. Optimise quantitative parameters such as concentrations and volumes to grow better crystals. The proteins has to be pure, stable and soluble for up to 10mg/ml.
5. Peptide crystallization
Short peptides are crystallized via various evaporation techniques. Many peptides are difficult to form well-ordered crystals with uniform content due to their inherent flexibility. However once the crystals are formed, short peptides can be very stable and do not lose their structure and properties in dried conditions. The peptide purity is another important parameter. For most cases the peptide should be at least 95% or 98% pure. Presence of any contaminations can make crystals unsuitable for X-ray analysis. The selection of solvents is very important for peptide crystallization. From the peptide sequence, we should know if the peptide is hydrophilic, hydrophobic, aromatic, basic or acidic. Based on these properties, dissolve peptides at high concentration in candidate solvent such as 50% methanol or other organic solvents and set up crystallisation trials. After evaporation, the precipitated peptides in the reservoir can be in amorphous, crystalline or crystal state.
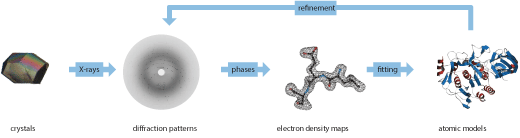
Structure Determination by X-Ray Crystallography
Protein structure can be determined by X-Ray crystallography and NMR. The steps in solving a protein crystal structure at high resolution are diagrammed above. The protein has to be first crystallized. Then the X-ray diffraction pattern including amplitudes and positions from the crystal are recorded. From these patterns, and with the use of phase information from labelled atoms in the crystal, electron density maps are computed. Lastly, a model of the protein is constructed, adjusted and refined from the electron density maps and the diffraction pattern.