Service Features
- For crude peptide libraries, we guarantee a purity >30%. Depending on the peptide sequence, crude purity can be as high as 80%
- For purified peptide libraries, we offer various purity levels: >70%, >80%, and up to 98%
- Peptide sequences may range in length from 3–20 amino acids
- The minimum number of peptides in each library ordered is 48
- Delivery in 2–3 weeks
- Certified analysis, HPLC, and MS reports are provided for each peptide
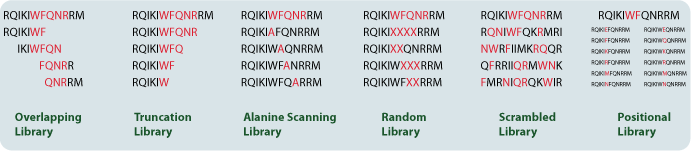
Library Types
Overlapping Peptide Library: Individual peptides can be divided in to several fragments that overlap. The resulting overlapping peptide libraries can then be used for processes including continuous and linear epitope mapping.
Truncation Peptide Library: Peptides can be cleaved systematically into small fragments. The library of truncated peptides could then be used to predict the minimum length amino acid that could be used to achieve optimal epitope activity.
Alanine Peptide Scanning Library: Each amino acid is substituted individually and systematically for alanine. Alanine scanning allows the easy identification of the specific amino acids that are responsible for the conformation, activity, and function of a protein.
Random Peptide Library: Selected positions are substituted with all 20 natural amino acids simultaneously, which might increase peptide activity.
Scrambled Peptide Library: These libraries are designed using variations of the original sequence of a peptide. The resulting peptides are used generally as negative controls to show that a specific sequence is critical to the protein function or activity. It is also a random screening tool used to find new leads.
Positional Peptide Library: Selected sites or regions within a peptide sequence are replaced systematically with other amino acids. These libraries facilitate the identification of the positions and regions that are responsible for specific activities or effects.
Applications
- Mapping protein-protein interaction sites: ELISAs and related assays, the immunoprecipitation-based identification of interacting proteins
- Antibody profiling: mapping and validating epitopes, the characterization of therapeutic antibodies, studying anti-antibody and neutralizing antibody actions in vitro. The overlapping peptides from LifeTein was used for epitope mapping. It was cited in Cell: Cell, Volume 159, Issue 4, 6 November 2014, Pages 844–856, DOI: 10.1016/j.cell.2014.10.032, A Noncanonical Frizzled2 Pathway Regulates Epithelial-Mesenchymal Transition and Metastasis
- Biomarker identification: studies related to cancer, and infectious and autoimmune diseases
- Mapping antigenic immunodominant regions: intracellular signaling pathways and protein fingerprinting
- The development of novel vaccines: peptide vaccines, peptide drug modifications, and vaccine efficacy testing
- Enzyme motif discovery and profiling: for enzymes including methyltransferases, kinases, phosphatases, proteases, and glycosyltransferases
Screening combinatorial peptide libraries to optimize enzyme substrates and create high-affinity protein ligands
Recent improvements in peptide delivery technologies coupled with data available from the human genome project have facilitated the identification of novel drug targets. This has also led to an increased demand for effective systems to create synthetic peptide libraries.
Synthetic peptide libraries could be used to screen a variety of molecules, such as enzymatic substrates and inhibitors, and cell-binding peptides. Combinatorial techniques, unlike traditional methodologies, allow many molecules to be handled simultaneously. Therefore, combinatorial chemistry is an important tool for discovering novel drug candidates, catalysts, and other related materials.
An important biological application of custom peptide libraries is characterization of the binding events that occur between specific proteins and their peptide ligands. Additional applications include identifying enzyme substrates, and characterizing the ligands that mediate cellular adhesion. For example, fluorescent protease substrates could be designed, placed in a peptide library, and used to screen protease substrates. Peptide libraries can also be used to quantify kinase activity, as well as a variety of detection methods including surface plasmon resonance (SPR), fluorescence, and phosphorimaging.
Peptide libraries can be used to rapidly identify high-affinity ligands when comparing the specificity of interactions among different domains.
Intracellular kinase-mediated signaling requires specific protein complexes, whose assembly is mediated and directed by phosphopeptide binding domains; proteins that bind to these phosphopeptides can be identified using peptide libraries. This allows proteins that bind specifically to phosphorylated peptide library affinity matrices, including pTyr, pSer/pThr-Pro, and pSer/pThr-pSer/pThr, to be identified.
For example, a phosphotyrosyl (pY) peptide library that contains completely random residues at different positions from pY could be screened against biotinylated domains, and the resulting beads bound to high-affinity ligands of the biotinylated domains could be identified using enzyme-linked assays.